Overview¶
This section describes how msys represents particles and forcefields, and the relationship between the msys representation and the various chemical file formats msys supports.
Molecular Structures¶
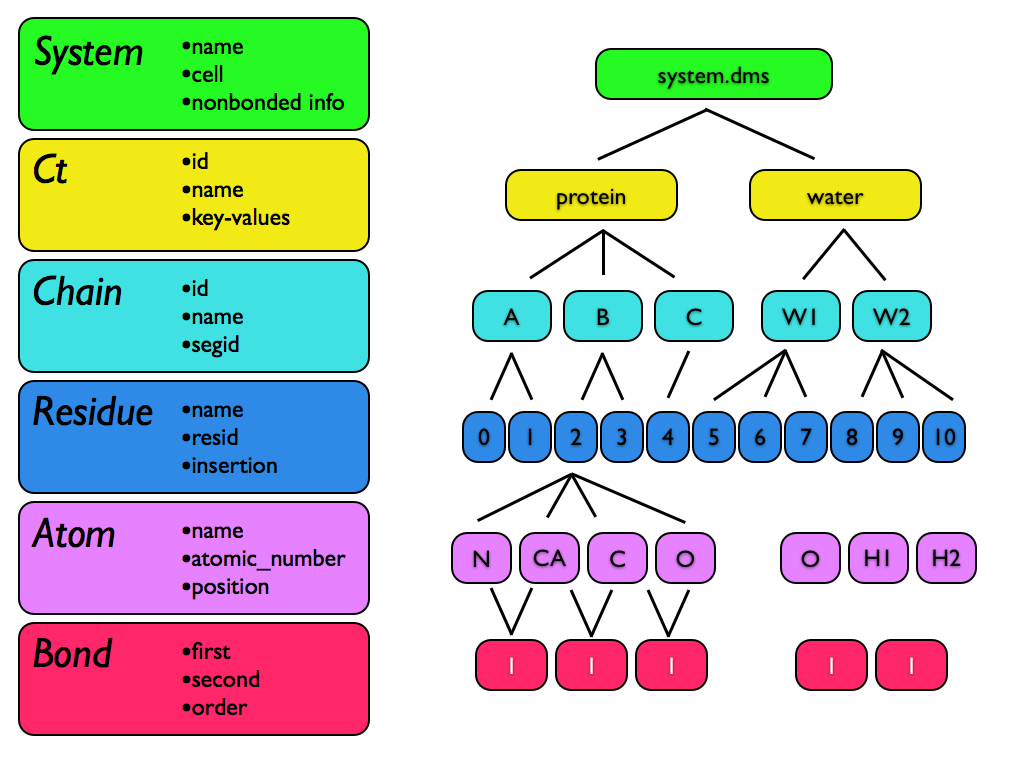
All molecular structure in Msys is held in an object called a System. Within a System, individual particles, including physical atoms as well as pseudo-particles used as interaction sites, are represented as Atoms. Bonds between Atoms are represented by a Bond structure. Atoms are grouped into Residues, and Residues are grouped into Chains. Finally, Chains are grouped into higher-level structures called “components”, or Cts for short.
Every structure type is contained within its parent type; thus, even a single particle System will contain (at least) one Residue, Chain, and Ct. If there is no information in the file to delineate separate Residues, Chains, etc., then only a single such entity will be created.
There are also several other miscellaneous tables in each System:
The cell holds the periodic cell information. It consists of three vectors, each with three components.
The nonbonded_info structure holds meta-information about the type of nonbonded interactions used in the forcefield.
There may be one or more auxiliary tables, indexed by name, which hold arbitrary additional forcefield data or other user-defined tables. These are indexed by name. The main use for auxiliary tables is to hold “cmap”-style tables from Charmm-style forcefields.
The Ct is the highest level of molecular organization, after the System. Many file formats, including MAE, SDF, etc., contain multiple structures, and it can be convenient to represent the entire contents of such a file in a single msys System without losing the distinction between structure records. When msys loads such a multi-component file, each entry gets placed in its own Ct. Another use for the Ct objects is when one System is appended to another. If there were no Ct objects, then Chains in one system might be unintentionally combined with Chains in the other system if the Chains had the same name. Finally, Ct blocks provide a space for arbitrary metadata about system components to be stored.
Chains in msys represent collections of Residues. Their main purpose is to hold the traditional chain and segment name information used in popular formats such as PDB.
Chains have just two settable properties: name and segid. When loading chemical systems, Residues are grouped into Chains entities based on their chain name and/or segid in the file, whichever is applicable.
A Residue in msys is a collection of Atoms. Residues have three settable attributes: name, resid, and insertion.
Finally, the Atom class represents all particles in the System, including real atoms as well as virtual and dummy particles. Each Atom has an atomic number, position, mass, and a number of other built-in properties.
Forcefields¶
A System also holds a set of TermTables representing the interactions
between Atoms. A TermTable can be thought of as a particular kind
of interaction; for example, a fully parameterized system would likely
contain a stretch_harm TermTable to represent two-body covalent
bond forces. Each Term in a TermTable refers to the same number
of atoms, though there can be any number of Terms in a given TermTable.
Typically, many of the interactions in a TermTable are parameterized using identical parameters, especially when there are many identical copies of the same molecule in the System. For compactness, and also for ease of forcefield parameterization, a TermTable holds a separate table called a ParamTable which contains the interaction properties that can be shared by many Terms. Changes to an entry in a ParamTable will affect the interaction strengths of every Term referencing that entry. However, as illustrated below, operations on an individual Term will affect the interaction properties of just that Term; behind the scenes, Msys takes care of creating a copy of a Term’s parameters as needed.
It is also possible for developers to construct multiple TermTables that share the very same ParamTable, so that changes to a shared ParamTable affect multiple TermTables or Systems.
Reading and Writing Files¶
Msys reads and writes many popular chemical file formats. While most file formats have some concept of particles, residues, and chains, the way in which these groupings are specified varies by file type. Even within a file type, groupings are not always done consistently; for example, a PDB file might have both segment and chain identifiers, and there is no requirement in the file that there be any relationship between them.
In addition, many chemical file formats, including MAE, MOL2, SDF, as well as DMS, can contain multiple, logically distinct chemical groups or components. In some contexts, such as an MD simulation, it makes sense to consider all the components as part of a single system. In other contexts, such as processing a large batch of ligand structures, one wants to consider the components one at a time.
Forcefield information is also present in different file types in widely disparate forms. If forcefield information is read in one format and written out in another, it must be done with minimal loss of precision.
Mapping of residues and chains¶
As mentioned earlier, Msys groups all Atoms into Residues, and all Residues into Chains. This hierarchy is, unfortunately, rarely made explicit in the chemical system files in wide use, so Msys must infer the grouping based on the values of certain particle attributes.
Msys uses the chain and segid particle properties to group Residues
into Chains. Within a chain, Atoms are grouped into Residues based
on their resname and resid attributes. Thus, in Msys, every Atom
within a given Residue has by definition the same resname and resid.
By the same token, every Atom and Residue within a given Chain has
the same chain and segid.
Upon loading a system, the number of Chains will be given by the number
of distinct chain and segid pairs appearing in the particle table,
and, within a given Chain, the number of Residues will be given by
the number of distinct resname and resid pairs appearing in atoms
sharing the Chain’s chain and segid. After loading a system,
one is free to modify the resname and resid of any Residue.
Bear in mind, however, that if two initially distinct Residues in the
same Chain come to have identical resname and resid, they will
be merged into a single Residue upon saving and loading.
Whitespace in atom, residue and chain names¶
The PDB file format specifies that atom and residue names should be
aligned to particular columns within a 4-column region. Unfortunately,
some have taken this alignment requirement to mean that an atom’s
name actually includes the surrounding whitespace! When Msys loads
a chemical system, the following fields are stripped of leading and
trailing whitespace before they are inserted into the structure: name
(atom name), resname (residue name), chain (chain identifier),
and segid (segment identifier).